Research
Members of the McTavish Lab focus on developing efficient methods that take advantage of the novel information in large scale data sets, with an emphasis on reproducibility and open science.
Current projects:
-
Extensiphy: Placing taxa in a sequence alignment and phylogeny using Next Generation Sequencing reads
Testing the effects of choice of reference genome and SNP calling pipeline parameters on phylogenetic inference.
-
TreeToReads: Simulating pipelines to generate Next Generation Sequencing reads from realistic phylogenies
-
Physcraper: Automated phylogenetic updating
Physcraper GitHub repo Documentation Examples

-
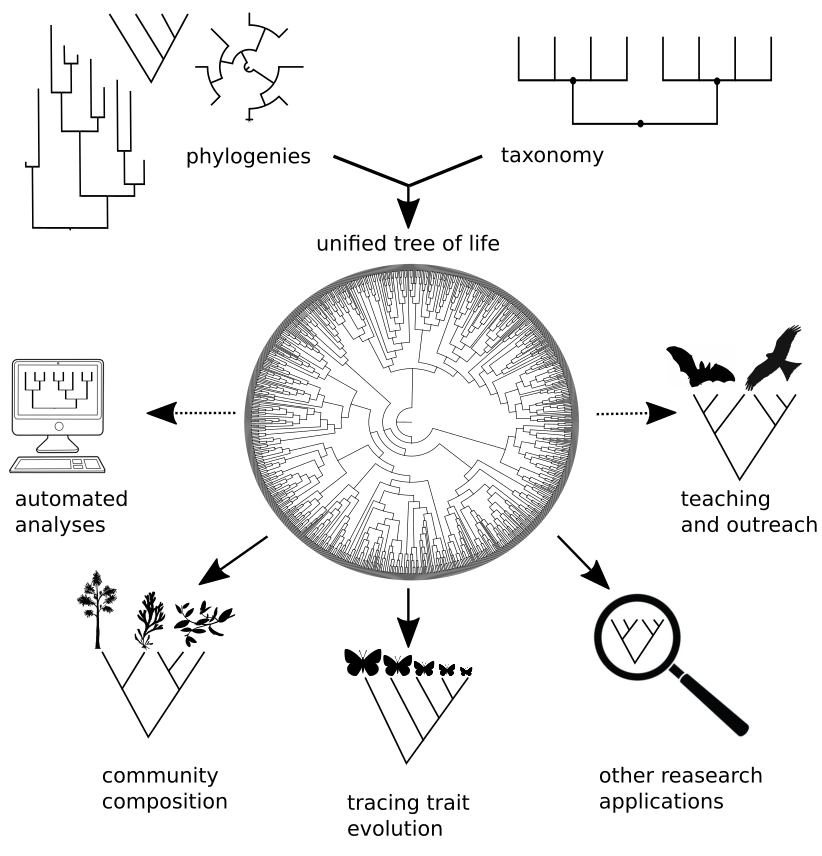
The Open Tree of Life
A synthetic phylogenetic tree of all life.

UC Merced writup of recent Open Tree of life grant