Getting a piece of the Synthetic Open Tree of Life
Overview
Teaching: 5 min
Exercises: 5 minQuestions
What is the synthetic Open Tree of Life?
How do I interact with it?
Why is my taxon not in the tree?
Objectives
Get an induced subtree
Get a subtree
The synthetic Open Tree of Life (synthetic OpenTree from now on) summarizes information from 1239 trees from 1184 peer-reviewed and published studies, that have been uploaded to the OpenTree database (the Phylesystem) through a curator system.
Functions from the rotl package that interact with the synthetic OpenTree start with tol_.
To access general information about the current synthetic OpenTree, we can use the function tol_about(). This function requires no argument.
rotl::tol_about()
OpenTree Synthetic Tree of Life.
Tree version: opentree13.4
Taxonomy version: 3.3draft1
Constructed on: 2021-06-18 11:13:49
Number of terminal taxa: 2392042
Number of source trees: 1239
Number of source studies: 1184
Source list present: false
Root taxon: cellular organisms
Root ott_id: 93302
Root node_id: ott93302
This is nice!
As you can note, the current synthetic OpenTree was created not too long ago, on 2021-06-18 11:13:49.
This is also telling us that there are currently more than 2 million tips on the synthetic OpenTree.
It is indeed a large tree. So, what if we just want a small piece of the whole synthetic OpenTree?
Well, now that we have some interesting taxon OTT ids, we can easily do this.
Getting an induced subtree
The function tol_induced_subtree() allows us to get a tree of taxa from different taxonomic ranks.
resolved_names$ott_id
my_tree <- rotl::tol_induced_subtree(ott_ids = resolved_names$ott_id)
Warning in collapse_singles(tr, show_progress): Dropping singleton nodes
with labels: Mammalia ott244265, Theria (subclass in Deuterostomia)
ott229558, Eutheria (in Deuterostomia) ott683263, Boreoeutheria ott5334778,
Laurasiatheria ott392223, mrcaott1548ott6790, mrcaott1548ott3607484,
mrcaott1548ott4942380, mrcaott1548ott4942547, mrcaott1548ott3021, Artiodactyla
ott622916, mrcaott1548ott21987, mrcaott1548ott5256, mrcaott5256ott4944931,
Whippomorpha ott7655791, Cetacea ott698424, mrcaott5256ott3615450,
mrcaott5256ott44568, Odontoceti ott698417, mrcaott5256ott5269,
mrcaott5269ott6470, mrcaott5269ott47843, mrcaott47843ott194312,
mrcaott4697ott263949, Carnivora ott44565, Caniformia ott827263,
Canidae ott770319, mrcaott47497ott3612617, mrcaott47497ott3612529,
mrcaott47497ott3612596, mrcaott47497ott3612516, mrcaott47497ott3612589,
mrcaott47497ott3612591, mrcaott47497ott3612592, mrcaott47497ott77889,
Feliformia ott827259, mrcaott6940ott19397, mrcaott19397ott194349, Felidae
ott563159, mrcaott54737ott660452, mrcaott54737ott86170, mrcaott54737ott86175,
mrcaott54737ott442049, mrcaott54737ott86162, mrcaott54737ott86166, Sauropsida
ott639642, Sauria ott329823, mrcaott246ott4128455, mrcaott246ott4127082,
mrcaott246ott4129629, mrcaott246ott4142716, mrcaott246ott4126667,
mrcaott246ott1662, mrcaott246ott2982, mrcaott246ott31216, mrcaott246ott4947920,
mrcaott246ott4127428, mrcaott246ott4126230, mrcaott246ott4127421,
mrcaott246ott664349, mrcaott246ott4126505, mrcaott246ott4127015,
mrcaott246ott4129653, mrcaott246ott4127541, mrcaott246ott4946623,
mrcaott246ott4126482, mrcaott246ott4128105, mrcaott246ott4127288,
mrcaott246ott4132146, mrcaott246ott3602822, mrcaott246ott4143599,
mrcaott246ott3600976, mrcaott246ott4132107, Aves ott81461, Neognathae
ott241846, mrcaott246ott5481, mrcaott246ott5021, mrcaott246ott7145,
mrcaott246ott5272, mrcaott5272ott9830, mrcaott9830ott86672, mrcaott9830ott90560,
mrcaott9830ott18206, mrcaott18206ott60413, Sphenisciformes ott494366
Note: What does this warning mean?
This warning has to do with the way the synthetic OpenTree is generated. You can look at the overview of the synthesis algorithm for more information.
Let’s look at the output of tol_induced_subtree().
my_tree
Phylogenetic tree with 5 tips and 4 internal nodes.
Tip labels:
Delphinidae_ott698406, mrcaott47497ott110766, Felis_ott563165, Spheniscidae_ott494367, Amphibia_ott544595
Node labels:
Tetrapoda ott229562, Amniota ott229560, mrcaott1548ott4697, mrcaott4697ott6940
Rooted; no branch lengths.
R is telling us that we have a rooted tree with no branch lengths and 5 tips. If we check the class of the output, we will verify that it is a ‘phylo’ object.
class(my_tree)
[1] "phylo"
A ‘phylo’ object is a data structure that stores the necessary information to build a tree.
There are several functions from different packages to plot trees or ‘phylo’ objects in R (e.g., phytools). For now, we will use the one from the legendary ape package plot.phylo():
ape::plot.phylo(my_tree, cex = 2) # or just plot(my_tree, cex = 2)
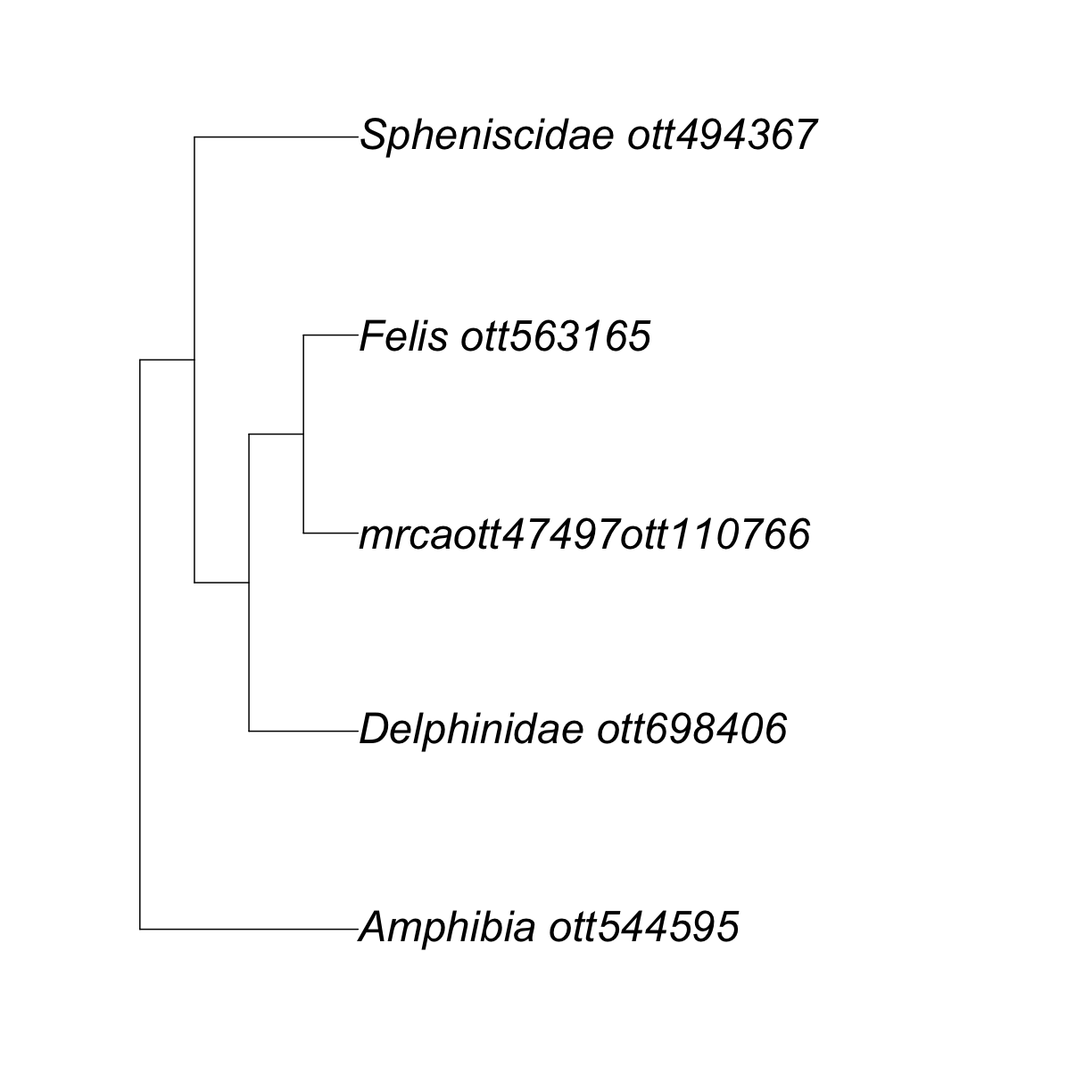
This is cool!
But, why oh why did my Canis disappear? 😢
Well, it did not actually disappear, it was replaced by the label “mrcaott47497ott110766”.
We will explain why this happens in the next section.
Now, what if you want a piece of the synthetic OpenTree containing all descendants of your taxa of interest?
Getting a subtree of one taxon
We can extract a subtree of all descendants of one taxon at a time using the function tol_subtree() and an OTT id of your choosing.
Let’s extract a subtree of all amphibians.
First, get its OTT id. It is already stored in our resolved_names object:
amphibia_ott_id <- resolved_names["Amphibia",]$ott_id
Or, you can run the function tnrs_match_names() again if you want.
amphibia_ott_id <- rotl::tnrs_match_names("amphibians")$ott_id
Now, extract the subtree from the synthetic OpenTree using tol_subtree().
amphibia_subtree <- rotl::tol_subtree(ott_id = resolved_names["Amphibia",]$ott_id)
Let’s look at the output:
amphibia_subtree
Phylogenetic tree with 10020 tips and 4669 internal nodes.
Tip labels:
Odorrana_geminata_ott114, Odorrana_chapaensis_ott214633, Odorrana_grahami_ott43280, Odorrana_margaretae_ott440550, Odorrana_kuangwuensis_ott3618367, Odorrana_junlianensis_ott656728, ...
Node labels:
Amphibia ott544595, Batrachia ott471197, Anura ott991547, , , , ...
Unrooted; no branch lengths.
This is a large tree! We will have a hard time plotting it.
Now, let’s extract a subtree for the genus Canis. It should be way smaller!
subtree <- rotl::tol_subtree(resolved_names["Canis",]$ott_id)
Error: HTTP failure: 400
list(contesting_trees = list(`ot_278@tree1` = list(attachment_points = list(list(children_from_taxon = list("node242"), parent = "node241"), list(children_from_taxon = list("node244"), parent = "node243"), list(children_from_taxon = list("node262"), parent = "node255"), list(children_from_taxon = list("node270"), parent = "node267"))), `ot_328@tree1` = list(attachment_points = list(list(children_from_taxon = list("node519"), parent = "node518"), list(children_from_taxon = list("node523"), parent = "node522")))),
mrca = "mrcaott47497ott110766")[/v3/tree_of_life/subtree] Error: node_id was not found (broken taxon).
😱 😱 😱
What does this error mean??
A “broken” taxon error usually happens when phylogenetic information does not match taxonomic information.
For example, extinct lineages are sometimes phylogenetically included within a taxon but are taxonomically excluded, making the taxon appear as paraphyletic.
On the Open Tree of Life browser, we can still get to the subtree (check it out here).
From R, we will need to do something else first. We will get to that on the next episode.
Key Points
OTT ids and node ids allow us to interact with the synthetic OpenTree.
Portions of the synthetic OpenTree can be extracted from a single OTT id or from a bunch of OTT ids
It is not possible to get a subtree from an OTT id that is not in the synthetic tree.