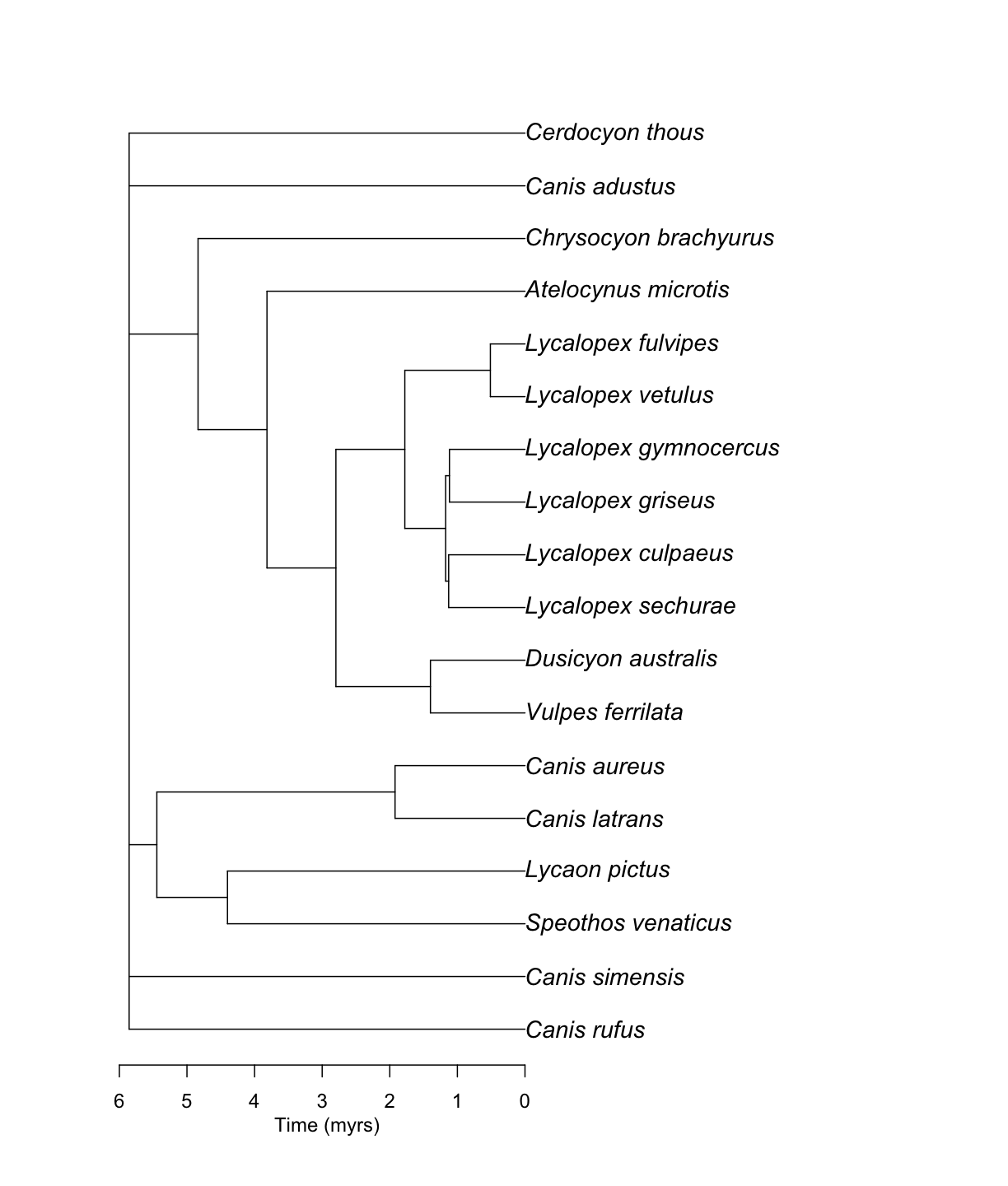
Summarizing branch length information
Overview
Teaching: 5 min
Exercises: 5 minQuestions
How do I summarize information from different source chronograms?
How do I choose a preferred source chronogram?
Objectives
Understanding the depth of uncertainty around age estimates.
Now that we have a collection of chronograms containing our taxa of interest, we can go on to summarize the information in them.
There is no consensus on the best way to do this.
We have implemented two ways of summarizing information from several chronograms into a single one. The fastest one is using the median of node ages for each node with available information, and then evenly distributing ages across nodes.
canis_phylo_median <- datelife::summarize_datelife_result(canis_dr, summary_format = "phylo_median")
Check that we actually went from a list of matrices to a tree with branch lengths:
canis_phylo_median
Phylogenetic tree with 18 tips and 13 internal nodes.
Tip labels:
Canis_rufus, Canis_simensis, Speothos_venaticus, Lycaon_pictus, Canis_latrans, Canis_aureus, ...
Node labels:
n1, n2, n3, n4, n5, n6, ...
Unrooted; includes branch lengths.
Good. Now we can plot our chronogram!
ape::plot.phylo(canis_phylo_median, cex = 1.2)
# Add the time axis:
ape::axisPhylo()
# And a little hack to add the axis name:
graphics::mtext("Time (myrs)", side = 1, line = 2, at = max(get("last_plot.phylo",envir = .PlotPhyloEnv)$xx) * 0.5)
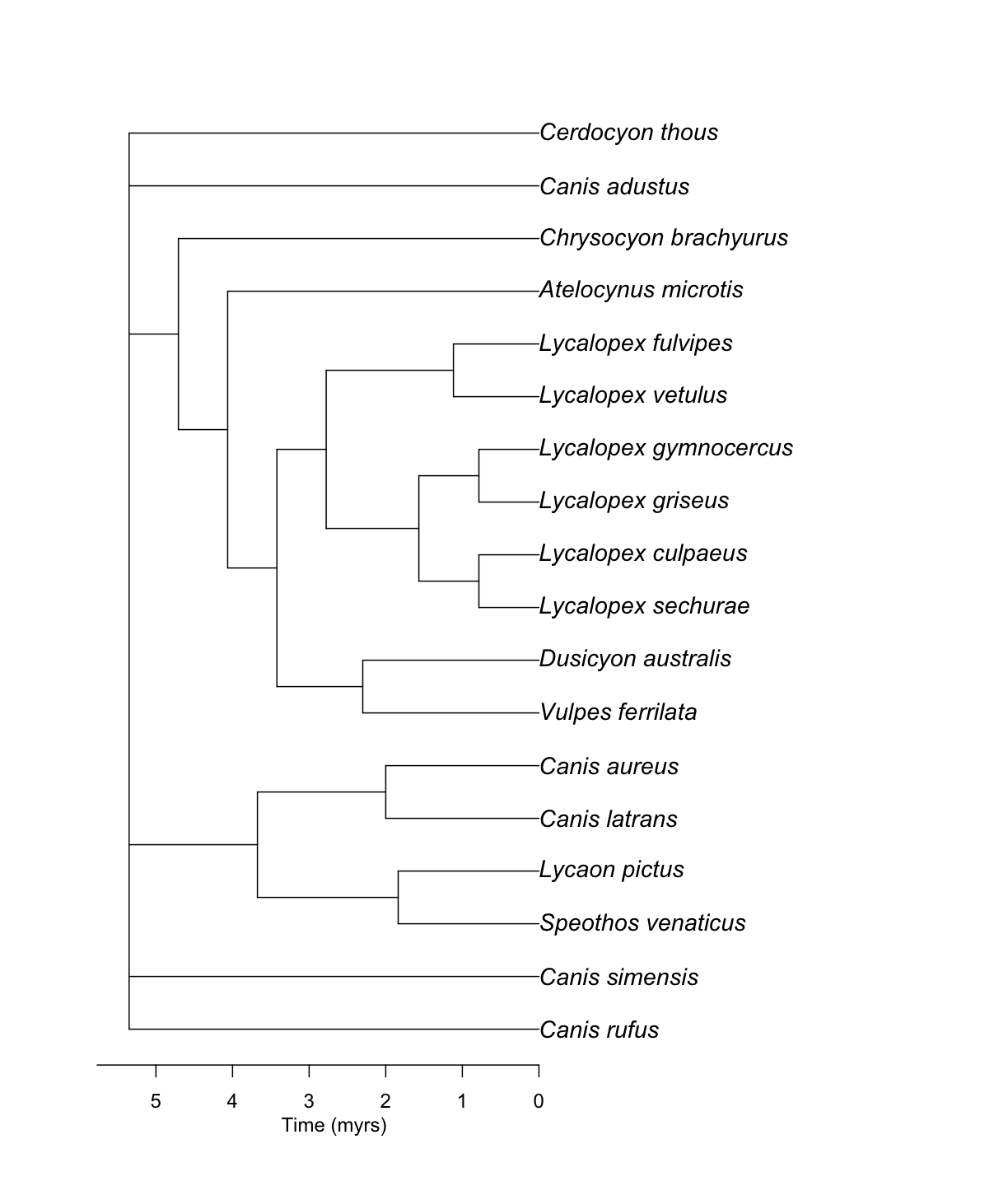
Challenge! Get the other type of summary chronogram
Hint: Explore options from the argument summary_format in the function
summarize_datelife_result()Solution
canis_phylo_sdm <- datelife::summarize_datelife_result(canis_dr, summary_format = "phylo_sdm")canis_phylo_sdmPhylogenetic tree with 18 tips and 13 internal nodes. Tip labels: Canis_rufus, Canis_simensis, Speothos_venaticus, Lycaon_pictus, Canis_latrans, Canis_aureus, ... Node labels: n1, n2, n3, n4, n5, n6, ... Unrooted; includes branch lengths.ape::plot.phylo(canis_phylo_sdm, cex = 1.2) ape::axisPhylo() graphics::mtext("Time (myrs)", side = 1, line = 2, at = max(get("last_plot.phylo",envir = .PlotPhyloEnv)$xx) * 0.5)
As you can note, the SDM sumary chronogram is slightly older than the median summary chronogram!
Finally, give it a try on the web browser of datelife, too. You can do the same things using a graphical user interface. It is fun!
Key Points
Source chronograms have a wide range of variation in age estimates.