Getting an induced subtree of all taxa within a taxonomic rank
Overview
Teaching: 5 min
Exercises: 5 minQuestions
How do I get all taxa from a certain taxonomic rank?
Objectives
Get an induced subtree from all taxa of a given taxonomic rank.
There is not a specific function in the package rotl that gets all taxa from a given taxonomic rank.
We will now shift to the datelife package and use the get_ott_children() function, that extracts OTT ids of all taxa from a rank specified by the argument ott_rank.
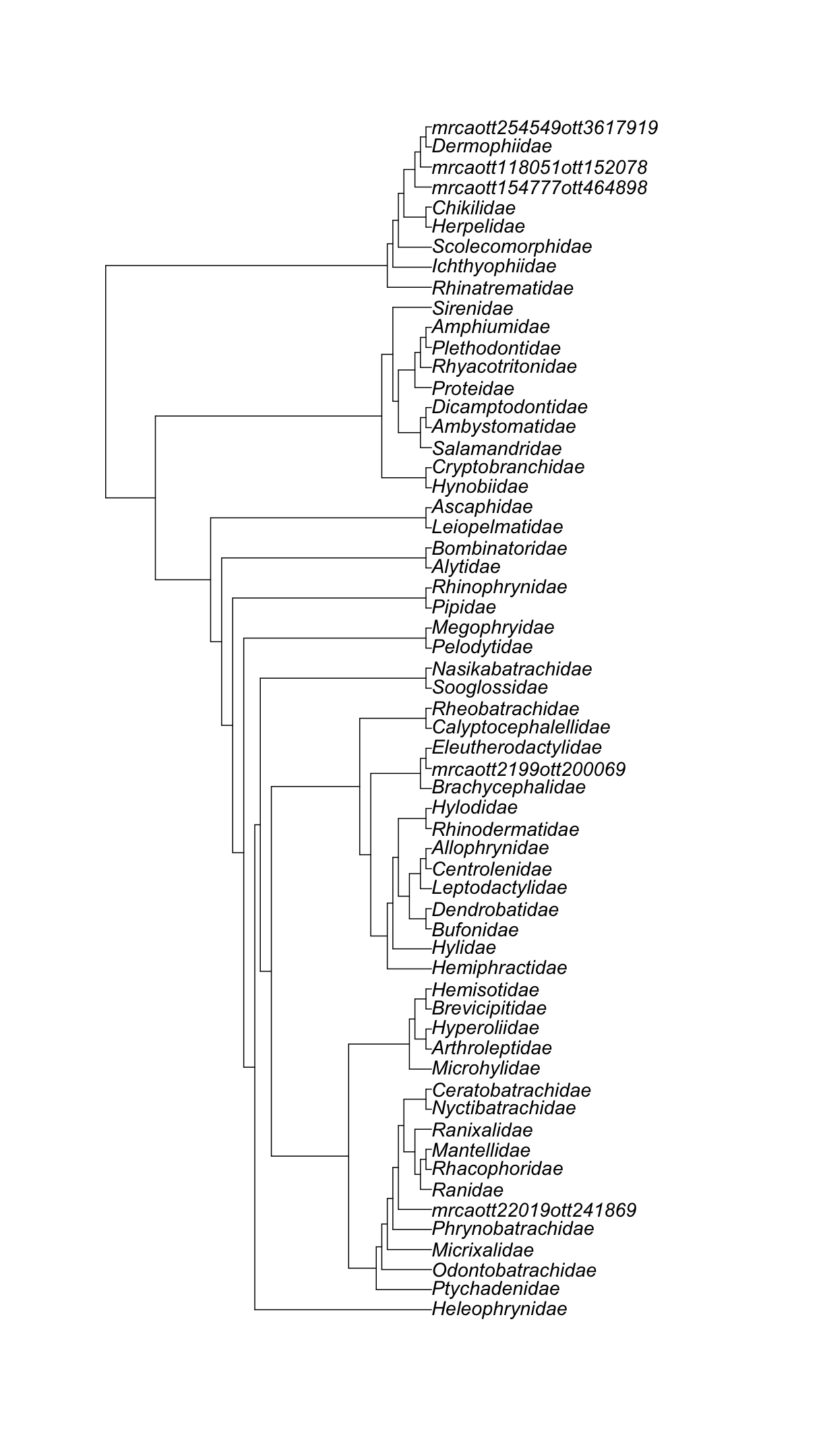
Let’s get all amphibian families.
amphibia_families <- datelife::get_ott_children(ott_ids = resolved_names["Amphibia",]$ott_id, ott_rank = "family")
str(amphibia_families)
List of 1
$ Amphibia:'data.frame': 70 obs. of 2 variables:
..$ ott_id: int [1:70] 118029 639647 639653 654645 128153 114139 114359 861429 379929 4948197 ...
..$ rank : chr [1:70] "family" "family" "family" "family" ...
Now, get the induced subtree using the amphibian families’ OTT ids.
amphibia_families_subtree <- rotl::tol_induced_subtree(amphibia_families$Amphibia$ott_id)
amphibia_families_subtree
Phylogenetic tree with 60 tips and 59 internal nodes.
Tip labels:
Ranidae_ott364560, Rhacophoridae_ott432783, Mantellidae_ott38969, Ranixalidae_ott403946, Nyctibatrachidae_ott1081210, Ceratobatrachidae_ott1081207, ...
Node labels:
Amphibia ott544595, Batrachia ott471197, Anura ott991547, mrcaott114ott3129, mrcaott114ott37876, mrcaott114ott18818, ...
Rooted; no branch lengths.
Let’s print the output.
ape::plot.phylo(amphibia_families_subtree, cex = 1.2)
Super cool!
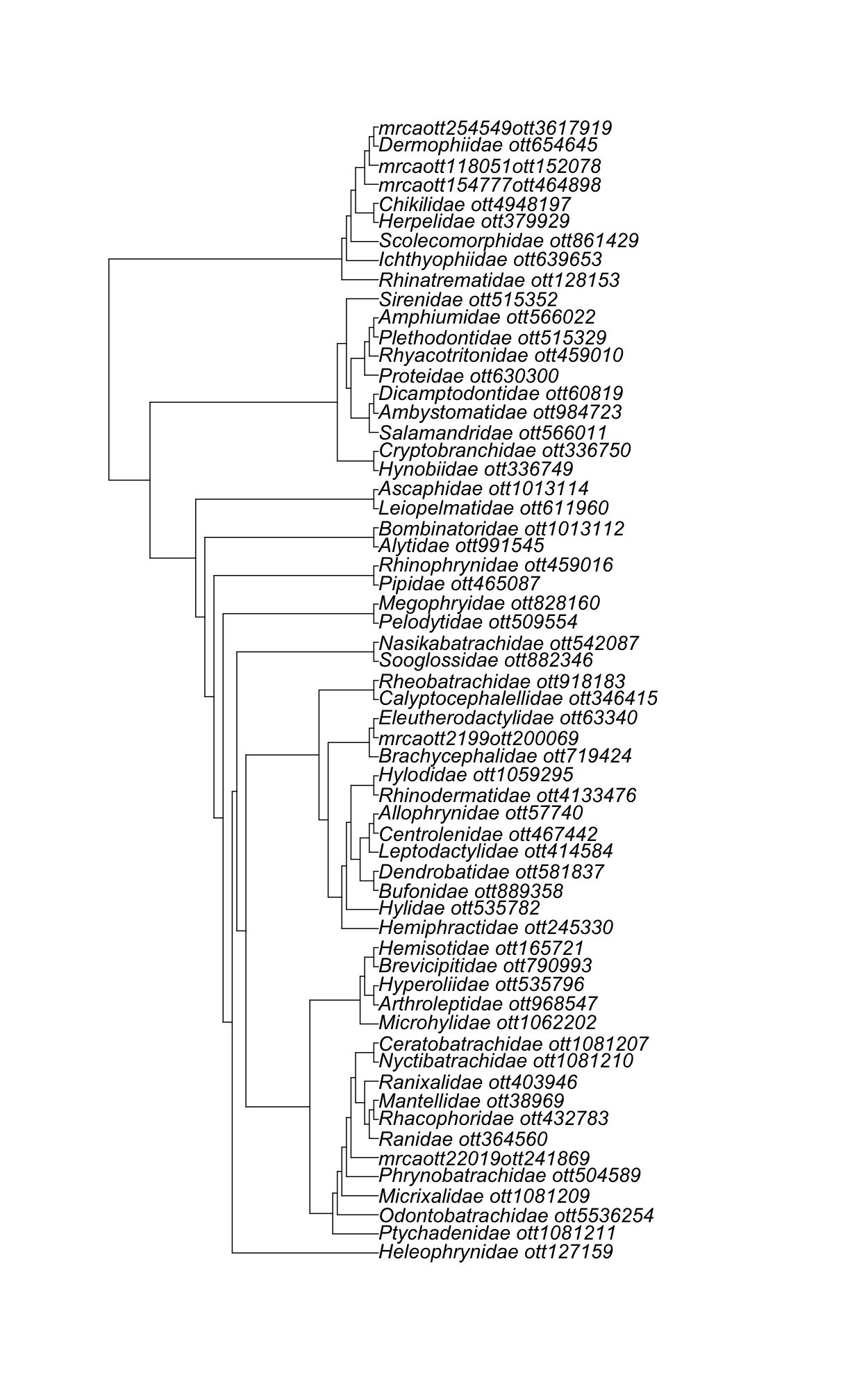
Hands on! Get a family subtree without ott ids in the tip labels
Hint: Look at the arguments of function
tol_induced_subtree()Solution
amphibia_families_subtree2 <- rotl::tol_induced_subtree(amphibia_families$Amphibia$ott_id, label_format = "name")Warning in collapse_singles(tr, show_progress): Dropping singleton nodes with labels: mrcaott114ott391676, mrcaott15857ott152667, mrcaott270630ott3618180, mrcaott22583ott100573, mrcaott22583ott44382, mrcaott44382ott72638, mrcaott44382ott100564, mrcaott65695ott254163, mrcaott65695ott121259, mrcaott2199ott411156, mrcaott7464ott21502, mrcaott21502ott918196, Pelobatoidea, mrcaott18818ott47772, Sirenoideaape::plot.phylo(amphibia_families_subtree2, cex = 1.2)
We have seen up to now how to get a portion of the synthetic OpenTree. How do I inspect the source phylogenetic trees that support the subtrees?
Pro Tip 4.1: Get all taxa from a taxonomic rank.
While
datelifefacilitates this task, there are other ways to get all taxa from a taxonmic rank using mostlyrotlfunctions. Try it out!amphibia_taxonomy <- rotl::taxonomy_subtree(resolved_names["Amphibia",]$ott_id[[1]]) ls(amphibia_taxonomy) length(amphibia_taxonomy$tip_label) head(amphibia_taxonomy$tip_label) tail(amphibia_taxonomy$tip_label) amphibia_taxonomy$edge_label edges <- datelife::extract_ott_ids(x=amphibia_taxonomy$edge_label) length(edges) # The following line takes a while to run! edges_taxon_info <- rotl::taxonomy_taxon_info(edges) ls(edges_taxon_info[[1]]) is_family <- unname(unlist(sapply(edges_taxon_info, "[", "rank") %in% "family")) is_suppressed <- unname(unlist(sapply(edges_taxon_info, "[", "is_suppressed_from_synth"))) # flag "is suppressed from synth" is not updated, so it is useless for now. amphibia_families <- unname(unlist(sapply(edges_taxon_info, "[", "ott_id")[is_family])) in_tree <- rotl::is_in_tree(amphibia_families) amphibia_families_subtree <- rotl::tol_induced_subtree(amphibia_families[in_tree])
Key Points
It is possible to get all types of subsets from the synthetic tree, as long as you can get the OTT ids!